
Abstract
Galaxy formation is a complex problem that connects large scale cosmology with small scale astrophysics over cosmic timescales. Hydrodynamical simulations are the most principled approach to model galaxy formation, but have large computational costs. Recently, emulation techniques based on Convolutional Neural Networks (CNNs) have been proposed to predict baryonic properties directly from dark matter simulations. The advantage of these emulators is their ability to capture relevant correlations, but at a fraction of the computational cost compared to simulations. However, training basic CNNs over large redshift ranges is challenging, due to the increasing non-linear interplay between dark matter and baryons paired with the memory inefficiency of CNNs. This work introduces EMBER-2, an improved version of the EMBER (EMulating Baryonic EnRichment) framework, to simultaneously emulate multiple baryon channels including gas density, velocity, temperature and HI density over a large redshift range, from z=6 to z=0. EMBER-2 incorporates a context-based styling network paired with Modulated Convolutions for fast, accurate and memory efficient emulation capable of interpolating the entire redshift range with a single CNN. Although EMBER-2 uses fewer than 1/6 the number of trainable parameters than the previous version, the model improves in every tested summary metric including gas mass conservation and cross-correlation coefficients. The EMBER-2 framework builds the foundation to produce mock catalogues of field level data and derived summary statistics that can directly be incorporated in future analysis pipelines.
Architecture
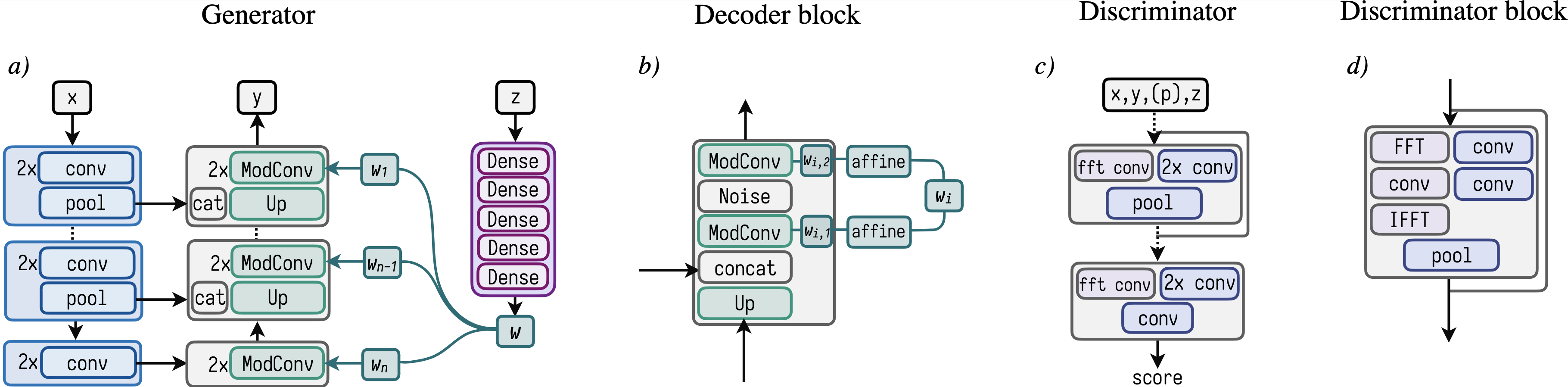
This figure depicts the main architecture design choices of the neural network model.
a)
Schematic overview of the generator based on the typical U-Net architecture. Features from the input x are extracted via an encoder (blue blocks) comprised of
two normal convolutions and a strided convolution for pooling. Subsequently the representations are decoded through a series of ModConv blocks to synthesize y.
The Mapping network shown in purple is a simple 5-layer multi-layer perceptron that maps the global redshift information z to the style vector w.
b)
Zoom-in of a single block in the decoder branch of the synthesis network comprised of a bicubic upsampling step, followed by a concatenation with the skip connection
from the corresponding encoder block. The base kernels of the two ModConvs are modulated via an affine layer with the externally mapped style vector w.
Between the two convolutions we concatenate 8 channels of gaussian noise to the tensor before feeding it through the second ModConv.
We emphasize that the two affine mappings are learned individually.
c)
Schematic of the discriminator comprised of individual blocks featuring a two-path strategy of convolutions in real and Fourier space.
The last convolution outputs a single scalar representing the score of the discriminator.
d)
Discriminator block comprised of two normal convolutions on the right path. Along the left path the data is first transformed to Fourier space,
convolved to extract frequency features and subsequently transformed back to real space via the inverse Fourier transform.
The data from the left and right path are added element-wise and pooled by a strided convolution.
Throughout all networks, we use kernel sizes of 3 and strides of 1 (except for the strided convolution, where a stride of 2 is used).
Movies
The following videos are part of the analysis performed on the learned distribution acting as a visual guide.
Citation
@article{bernardini_2025,
author = {Mauro Bernardini, Robert Feldmann, Jindra Gensior, Daniel Anglés-Alcázar, Luigi Bassini, Rebekka Bieri, Elia Cenci, Lucas Tortora and Claude-André Faucher-Giguère},
title = {EMBER-2: Emulating baryons from dark matter across cosmic time with deep modulation networks},
journal = {Monthly Notices of the Royal Astronomical Society},
doi = {10.1093/mnras/staf341},
year = {2025}}